We are looking for a highly motivated employee as “studentische Hilfskraft” (HIWI) for our bioinformatic working group.
Time expenditure
- 10 hours/week
- Flexible working time / place
Tasks
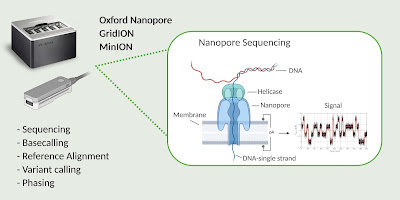
Nanopore sequencing is a sequencing method of the so-called third generation, which allows to sequence DNA and RNA directly, modification-sensitive and with unlimited length.
The development of new tools (command line) is rapid and one of your tasks will be to follow this development, test the latest tools and if possible, integrate them into our existing pipeline.
Requirements
You distinguish yourself through high reliability and organization and are able to work independently. You can be enthusiastic about new tasks. In addition to bioinformatics lectures, you have already been able to gain practical programming experience. Initial experience (or the willingness to quickly familiarize yourself) in the following areas is desirable:
- Linux command line, Bash, Python
- BLAST, Ensembl, IGV, alignment, variant calling
- Basic knowledge in genetics (SNPs, genes, genotypes, alleles)
Contact
Comments
Post a Comment